Our Research
New Method for Identifying Genetic Alterations that Modulate Gene Expression
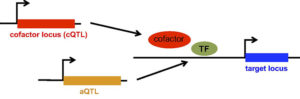
Read about our various methodologies for mapping trans-acting genetic determinants of gene expression in a recent news story on the website of the Department of Systems Biology.
Discovery of PQM-1 as a key regulator of aging and longevity
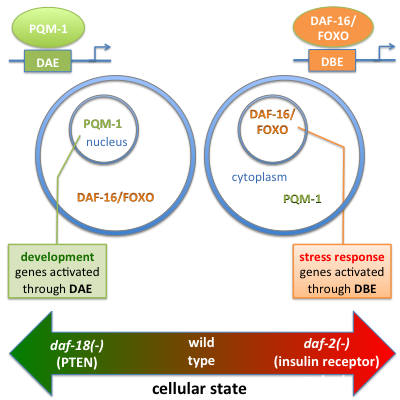
Aging is fundamental to the human life cycle and intimately connected with disease. Understanding the genetic and molecular determinants of animal longevity will provide new avenues for slowing the negative effects of aging. Using the nematode C. elegans as a model organism, and applying a combination of computational and experimental methodsin close collaboration with the lab of Dr. Coleen Murphy at Princeton University, Ronald Tepper in the Bussemaker lab has discovered (Tepper et al., Cell, 2013) that the little-studied transcription factor PQM-1 is a key regulator of development and longevity, and the long-sought factor that binds the so-called DAF-16 associated element (DAE). As it turns out, PQM-1 complements the well-known aging transcription factor DAF-16/FOXO in many respects. Both act as transcriptional activators, but they control distinct sets of target genes (stress response vs. growth). Whether DAF-16 or PQM-1 is nuclear or cytoplasmic depends on the status of the insulin/IGF-1 signaling pathway, but in opposite ways. This causes only one of the factors to be active as a regulator at any given time, depending on the conditions (e.g. low nutrients) and genetic background (e.g., loss of daf-2 or daf-18/PTEN). At the same time, as we have shown, the two factors interact with each other in an essential way: loss of pqm-1 affects DAF-16 subcellular localization, and vice versa. The molecular mechanisms underlying these important processes, however, remain obscure.
DSSR: New software for dissecting the spatial structure of RNA
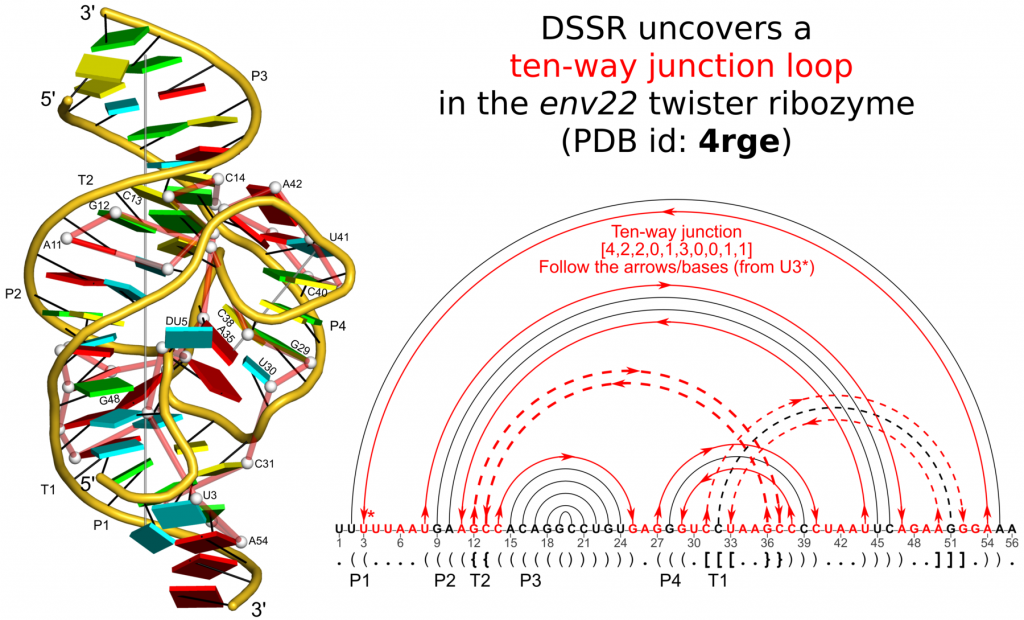
Insight into the three-dimensional architecture of RNA is essential for understanding its numerous cellular functions. Recent discoveries of new RNA folds and functions have stimulated interest in the development of technologies that can make sense of the complex spatial arrangements of these molecules. To address this need, Dr. Xiang-Jun Lu, an NIH-funded Associate Research Scientist affiliated with the Bussemaker Lab who previously developed the widely used 3DNAsoftware, has developed a new computational tool named DSSR (Dissecting the Spatial Structure of RNA). DSSR has already been integrated into Jmol and PyMOL, two widely used molecular graphics programs. It can serve as a cornerstone for RNA structural bioinformatics and will benefit a broad range of possible applications. [read the paper][download the software]